So, I downloaded the 20-year radar data set (Mar to Nov) from the German meteorological service (DWD). It contains about 1.5 million individual radar-derived precipitation fields – one every 5 minutes. And, I was very excited to run a rain cell tracking on it. After handing in my PhD thesis, I was going for a short trip to visit family and friends in Germany. Fortunately, I have a home server and I decided to let it do the heavy work while I am on holidays. It took the full ten days of my absence plus two extra days for the 8-core Intel CPU machine to process all data. So, let’s see how I did it.
This post is part of the germanRADARanalysis project.
The directory structure
In an earlier post, I showed how to download the radar data set, convert and restructure it as monthly NetCDF files. This makes it easier to process the data with my rain cell tracking algorithm. You can find the tracking software here on GitHub.
I created a new directory for this project (germanRADARanalysis/) and saved all NetCDF files in a sub-directory that I called RADARdata/. I then created the directory for the results of the rain cell tracking, celltracking/. The final directory structure looks like this.
germanRADARanalysis/
├── celltracking
└── RADARdata
├── raa01-yw2017.002_10000-200103-dwd---bin.nc
├── raa01-yw2017.002_10000-200104-dwd---bin.nc
├── raa01-yw2017.002_10000-200105-dwd---bin.nc
├── raa01-yw2017.002_10000-200106-dwd---bin.nc
├── raa01-yw2017.002_10000-200107-dwd---bin.nc
├── raa01-yw2017.002_10000-200108-dwd---bin.nc
├── ...Code language: Bash (bash)How the rain cell tracking works
I am using my own implementation of a rain cell tracking algorithm, celltrack. The concept for it was originally developed by Moseley et al. (2013). The most time consuming steps are the following:
- Search the precipitation field for continuous areas of rain (rain cells) and give them unique labels. A minimum threshold of 0.05 mm/5 minutes is applied to the precipitation field and cells smaller than 16 km2 are ignored.
- Look for overlaps between rain cells in subsequent time steps
- Calculate the velocity of each rain cell and average them on a relatively coarse grid of 3×4 tiles. This will give an estimation of the large-scale wind field.
- Advect all rain cells by the calculated large-scale wind field.
- Repeat steps 2 to 4 until the number of detected links converges. Six times is usually enough.
The final, less computationally expensive step is to connect all linked rain cells to tracks and label them. The following image shows an idealized version of an identified rain cell track.

A much more comprehensive explanation of the exact steps, definitions and available options of celltrack can be found in the documentation.
The radar data domain

The map above shows the location (red dots) of all available radar stations and the theoretical, maximal coverage (yellow area). The total extent of the data set lies within the green grid area which indicates the tiling for the advection correction. The center of each tile is marked by a blue cross.
Choosing the right parameters
There is a number of parameters and options that determine the exact behavior of the algorithm. The table below gives an overview of the chosen options and values. However, the most important are the threshold and the minimum area for rain cells.
| Option(s) | Description | Value |
| -var <char> | Select the variable in the NetCDF file | precipitation |
| -thres <val> | The minimum value of precipitation. Grid points below this are set to zero. | 0.05 (mm/5minutes) |
| -minarea <val> | The minimum area of a rain cell. Smaller cells are ignored in all following steps. | 16 (km2) |
| -advcor -cx <val> -cy <val> | Activate and define the grid for the advection correction. The chosen values will split the domain into 3×4 tiles. See the figure above for a map. | 300 (cx) and 275 (cy) |
| -nadviter <val> | Number of iterations for advection correction. | 6 |
| -tstep <val> | This defines the time step of the NetCDF file. | 300 (seconds) |
| -maxv <val> | Allowed maximum velocity for rain cells to be considered for calculating the advection field. | 0.04 (km/s) |
| -nometamstr -metanc -tracknc | These options switch off/on certain output routines. | none |
Run the rain cell tracking
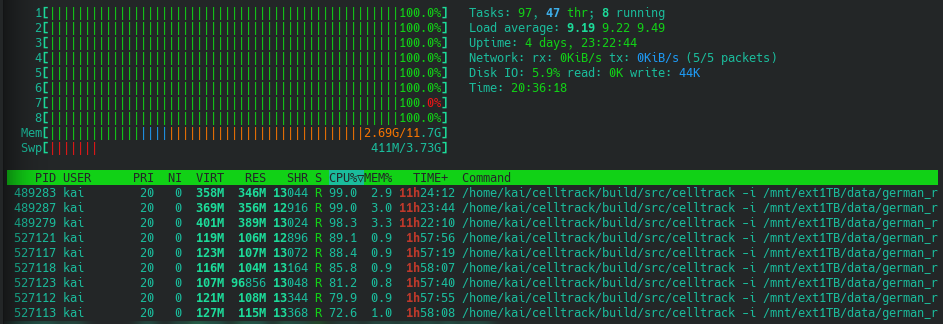
Once it was decided which options to use for the tracking, the execution was trivial. I created a script that uses xargs to parallelize the process to a good extent. A previous post of mine gives a detailed explanation of how this works. In a nutshell, the script starts eight instances of celltrack at a time. Each one for a different month of a specific year. As soon as all monthly files of a year are processed it steps to the next year and repeats the process. Here, I do not use xterm to show the process in individual terminal windows because I am running it on a headless server. Instead, each process writes its output to a text file.
#!/bin/bash
celltrack_bin=celltrack
project_dir=/home/kai/germanRADARanalysis/
output_dir=${project_dir}/celltracking/
data_dir=${project_dir}/RADARdata/
# first create the output directory structure
for year in $(seq -f "%04g" 2001 2020)
do
for month in $(seq -f "%02g" 3 11)
do
mkdir -p ${outpath}/${year}/${month}
done
done
# now run parallel instances of celltrack
for year in $(seq -f "%04g" 2001 2020)
do
printf '%s\n' "${months[@]}" | xargs -I "%" -P 8 /bin/bash -c "cd ${output_dir}/${year}/% && ${celltrack_bin} -i ${data_dir}/raa01-yw2017.002_10000-${year}%-dwd---bin.nc -var precipitation -thres 0.05 -advcor -tstep 300 -cx 300 -cy 275 -nadviter 6 -nometamstr -metanc -tracknc -maxv 0.04 -minarea 16 |& tee output_${year}%.txt"
done
exit 0Code language: Bash (bash)The script can also be downloaded from GitHub. When the script is finished (and it will take a while), the celltracking/ directory will have the following structure (directories only).
celltracking/
├── 2001
│ ├── 03
...
│ └── 11
├── 2002
│ ├── 03
...Each sub-directory contains the tracking results and the status output of celltrack.
Outlook
We have now a database of rain cell tracks for each month of the radar data set. In a future article, I will show how to handle the output and assemble it to a nicely structured data set with information about all identified rain cell tracks, such as the peak intensity, average intensity, area or even complete life cycles of tracks. We will also find out how to connect each track to a station record of dew point temperature and even the cloud type.